Detailed information
Genome browser
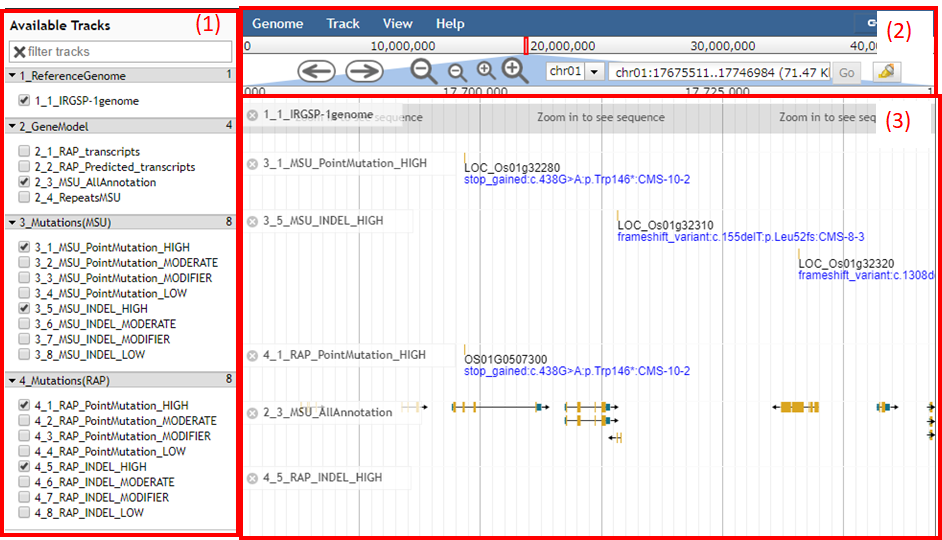
A Top screen of genome browser is separeted to three parts; (box 1) Selector of data tracks, (box 2) Selector of chromosomes and positions, and (box 3) viewer.
 Tweets by PGkyushu
Tweets by PGkyushu
Box 1
The box 1 contains the following data tracks selectors;
- 1_Reference genome The latest reference genome assembly of Oryza sativa ssp. japonica cv. Nipponbare, Os-Nipponbare-Reference-IRGSP-1.0 (Kawahara 2013).
- 1_1_1IRGSP-1.genome
- 2_GeneModel Two kind of major gene models, RAP model and MSU model, are available. Repeats database from MSU is also added to genome browser. If user has a request to integrate additional data tracks, please inform us.
- 2_1_RAP_transcript
- 2_2_RAP_Predicted_transcript
- 2_3_MSU_AllAnnotation
- 2_4_RepeatsMSU
- 3_Mutations(MSU) Mutations annotated by SnpEff's putative impact tags on MSU gene model. Detailed annotation description is cited from here.
- 3_1_MSU_PointMutation_HIGH
- 3_2_MSU_PointMutation_MODERATE
- 3_3_MSU_PointMutation_MODIFIER
- 3_4_MSU_PointMutation_LOW
- 3_5_MSU_INDEL_HIGH
- 3_6_MSU_INDEL_MODERATE
- 3_7_MSU_INDEL_MODIFIER
- 3_8_MSU_INDEL_LOW
- 4_Mutations(RAP) Mutations annotated by SnpEff's putative impact tags on RAP gene model. Detailed annotation description is cited from here.
- 4_1_RAP_PointMutation_HIGH
- 4_2_RAP_PointMutation_MODERATE
- 4_3_RAP_PointMutation_MODIFIER
- 4_4_RAP_PointMutation_LOW
- 4_5_RAP_INDEL_HIGH
- 4_6_RAP_INDEL_MODERATE
- 4_7_RAP_INDEL_MODIFIER
- 4_8_RAP_INDEL_LOW
Box 2
Navigator buttons are available to choose chromosomes, to move to left or right, and to zoom up and down genomic regions. User can specify chromosome and positions to be shown in genome browser with defined format (ex. chr01:17693661..17713500).
Or user can put gene ID in MSU and RAP. Several candidates are automatically suggested just a few letters are typed.


Box 3
Viewer demonstrates selected tracks of reference genome, gene models, and mutations.
The below figure is an example of mutations probably inducing premature stop codon at LOC_Os01g32280.
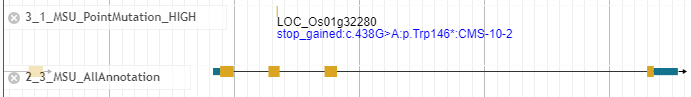
The mutation in above figure has description, "stop_gained:c.438G>A:p.Trp146*:CMS-10-2". The description represents that This mutation gains stop codon. G nucletide at 438th nucleotide changed to A nucleotide. Amino acid residue Trp changed to "*" stop codon. This mutation is found in M1 plant named "CMS-10-2".
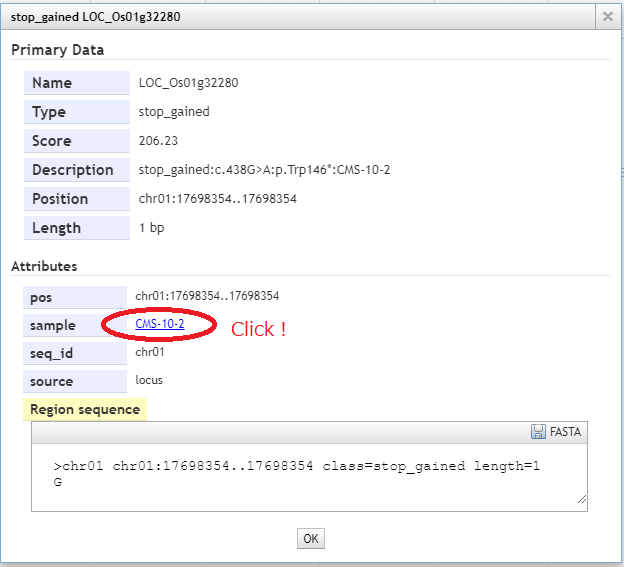
When the mutation is clicked, the popup window will be opened.The detailed informations is confirmed. When you clicked a sample term, a new genome browser specialized to the M1 plant "CMS-10-2" will be launched.
In the genome browser specified to the M1 plants "CMS-10-2", short read evidences covering mutation sites will be confirmed.
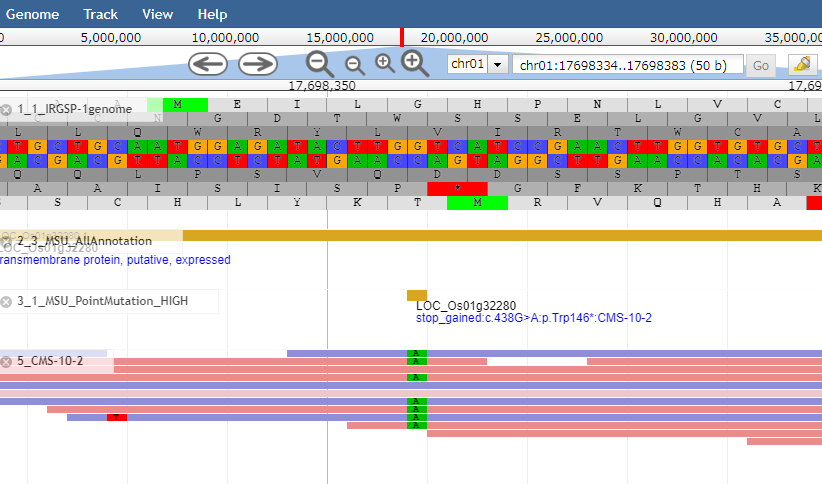
In the figure, 10 short reads were obtained at the mutation site. Of these, 8 reads carries nucleotide A (derived from allele with mutation) and the remaining 2 reads carries nucleotide G (derived from allele without mutation).
Work in table format (not genome browser)
Database construction
Chemical mutagenesis to fertilized eggs
We induced mutations to Oryza sativa ssp. japonica cv. Nipponbare (NB) by applying a chemical mutagens, N-methyl-N-nitrosourea (MNU) to fertilized egg cells at the single-cell stage in rice and achieved a high mutation frequency with MNU (Satoh and Omura 1979, 1986) due to a high mutation frequency by possible reduction or elimination of chimera formation and diplontic selection (competition of normal and mutant cells within the plant). The mutation rate was calculated as 7.4 × 10−6 per nucleotide, and the mutations were evenly distributed over the gene regions examined (Satoh et al. 2010). MNU is an SN1-type monofunctional alkylating agents with high reactivity towards oxygen atom sites of DNA, while SN2-type reagents such as EMS prefer to attack ring nitrogen atom sites of DNA. In particular, the MNU induces predominantly G to A (or C to T complementaryly) transitions. We obtain 1000 of M1 plants and their leaves are collected for whole genome sequencing.
Whole genome sequencing and Variant call
Whole genome sequencing were conducted on genomic DNA extracted from the M1 plants by Illumina short-read sequencing platform such as Hiseq X and Novaseq. Over 10 times coverage of shot reads were obtained per M1 plants.
Variant call analysis were conducted according to 'Best Practice' provided from Genome Analysis Tool Kit version 4 (GATK) (McKenna et al., 2010) with minor modifications. Briefly, short reads were mapped to reference genome assembly of NB, Os-Nipponbare-Reference-IRGSP-1.0 (Kawahara 2013), which is a unified release of the Rice Annotation Project (RAP) and the Michigan State University Rice Genome Annotation Project (MSU). After bam conversion and sorting by samtools (Li et al. 2009) and removal of PCR duplicate by Picard tool, high quality variants were submitted to base quality score recalibration (BQRS) by GATK to normalize base quality score of bam file. Nucleotide and indel variants were called by GATK GenotypeGVCFs tool.
Variant filtering
For variant filtering, we hypothesized that the same variant at the same site never occur among M1 plants. So the site where multiple M1 plans showed the same types of point mutations (PM) or Insertion/Deletion mutations (INDEL) were removed from the variant data for display in Genome browser in the MiRiQ database because these mutations were seemed to be sequencing error or mismapping of short reads possibly due to repetive sequences in genome. We did not apply any other filtering by quality score to screen variants as many as possible.
Mutation annotation by SnpEff
Gene annotation file (GFF) were downloaded from RAP-DB and MSU. Possible effects of mutations were analyzed by SnpEff (Cingolan et al. 2012). Suggested annotations with HIGH, MODERATE, MODIFIER, and LOW tags were separated and uploaded to Jbrowse Genome browser framework (Buels et al. 2016) for data demonstration
Statistics
Now on preparing.Access to the old versions
- Jbrowse of the MiRiQ0.0 (version 0.0 resulted from the 254 M1 plants).
- Jbrowse of the MiRiQ1.0 (version 1.0 resulted from the 721 M1 plants).
- Jbrowse of the MiRiQ2.0 (version 2.0 resulted from the 1154 M1 and M2 plants.
- Jbrowse of the MiRiQ3.0 (version 3.0 resulted from the 1354 M1 and M2 plants.
etc.
We expect MiRiQ is pronounced [miraikw].Reference
1. Kubo et al. (2022) Whole-Genome Sequencing of Rice Mutant Library Members Induced by N-Methyl-N-Nitrosourea Mutagenesis of Fertilized Egg Cell. Rice 15. https://doi.org/10.1186/s12284-022-00585-12. Kubo et al. (2024) MiRiQ Database: A Platform for In Silico Rice Mutant Screening. Plant Cell Physiol. 65:169-174. https://doi.org/10.1093/pcp/pcad134


